How To Register And Normalize Microscope Images
Abstract
Image registration of biological data is challenging as complex deformation problems are common. Possible deformation furnishings can be acquired in individual data preparation processes, involving morphological deformations, stain variations, stain artifacts, rotation, translation and missing tissues. The combining deformation effects tend to make existing automatic registration methods perform poor. In our experiments on series histopathological images, the 6 state of the fine art image registration techniques, including TrakEM2, SURF + affine transformation, UnwarpJ, bUnwarpJ, CLAHE + bUnwarpJ and BrainAligner, achieve no greater than 70% averaged accuracies, while the proposed method achieves 91.49% averaged accuracy. The proposed method has also been demonstrated to be significantly better in alignment of light amplification by stimulated emission of radiation scanning microscope brain images and serial ssTEM images than the criterion automatic approaches (p < 0.001). The contribution of this study is to introduce a fully automatic, robust and fast prototype registration method for 2nd image registration.
Introduction
Image registration is the procedure of transforming different sets of information into ane coordinate arrangement and elastic image registration is potentially an enabling technology for the constructive and efficient apply of many image-guided diagnostic and handling procedures, which rely on multimodality prototype fusion or series image comparison. The employ of elastic image registration covers a wide variety of medical applications, from edifice anatomical atlases, to longitudinal studies of tumor growth or other disease processes, through surgical planning and guidance1,2,iii,4. Consequently, information technology is a artistic field of research; techniques are numerous and inspired from a wide range of theories or techniques, such as statistics, data theory, theory of continuum mechanics, theory of thermodynamics, optical period, splines, wavelets and block matching and in that location are a number of reviews of biomedical image registration techniques and applicationsfive,6,7.
The histopathological study of tissue is an important tool in the medical field for the prognosis of disease. Conventionally, histological slides are examined nether an optical microscopy to reveal two dimensional images. However, this method is bereft to analyze circuitous 3 dimensional histology of lesion or tumor. By registering consecutive slices of biological images, 3D histology reconstruction can exist generated8. In 2010, Peng et al.9 introduced a Vaa3D system for real-time visualization of a three-dimensional digital atlas of neurite tracts in the fruitfly encephalon. Another useful application is multimodal molecular mapping, requiring the joint analysis of two-dimensional gene and poly peptide expression maps (obtained from in situ hybridization and immunohistochemistry, respectively) and/or their local comparison against cellular morphology maps (from histology)10. 3D reconstruction and visualization of microscopic images makes it possible to provide medical doctors and biologists a rapid and precise data prepare about the patients or animals regarding the micro-environment and topology in the tissue or organ potentially involved in certain diseases or biological functions.
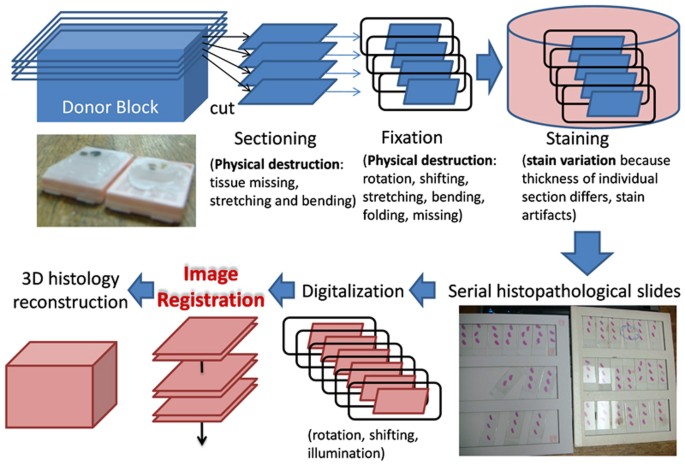
Serial slides can exist manually aligned by setting up a number of pairs of respective control points to the aforementioned (ten, y) location for consecutive images z i and z i +1 and the pairs of images and paired-sets of control points are then given to semi-automatic softwareeleven for image alignment. Fully automated registration of biological images is possible as demonstrated by software TrakEM212,13,fourteen,fifteen and in various studies8,10,xvi,17,18. All the same, 3D reconstruction of histopathological data is challenging. Cardona et al.12 pointed out that "TrakEM2 acknowledges that whatever automatic procedure (such as image registration and image segmentation) will somewhen fail partially or fully and volition require manual correction past a human being operator". In comparison to the CT scans or confocal images as used in the studies9,18 where the serial paradigm data maintains the property of geometrical continuity in 3d space, there are complex deformation problems for serial histopathological slides, including concrete destructions caused by cut and fixation, staining artifacts and uneven stain variations due to potential discrepancy in thickness of individual tissue sections. These complex baloney effects makes prototype registration of histological data a difficult chore (see Figure ane). In previous studies8,x,fourteen,15,sixteen,17,nineteen, the raw data tends to exist advisedly prepared, showing little morphological distortions or stain variation, to simplify the registration job. In improver, they tend to work with depression resolution images without dealing with high precision tissue or cell level registration. Effigy 2 illustrates possible deformation problems caused in individual information preparation processes, involving morphological deformations, stain variations, stain artifacts, rotation, translation and missing tissues. The combining deformation effects tend to make existing automatic registration methods perform poor.
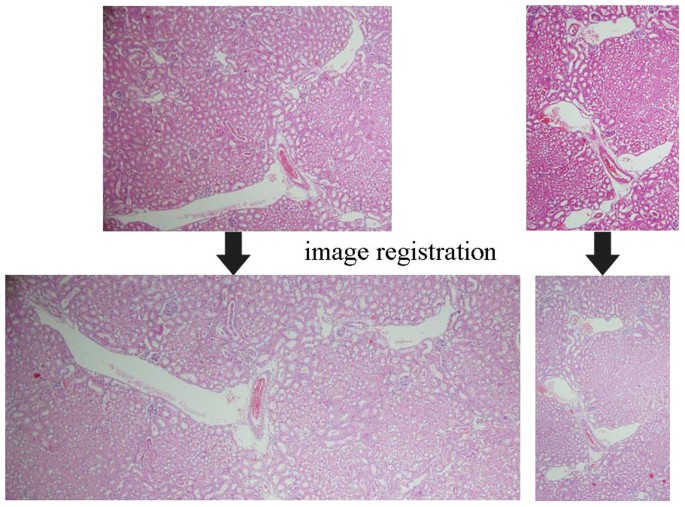
Image registration of histopathological images is difficult.
Paradigm registration is the process of transforming different sets of data into ane coordinate system, just for registering histopathological images, in that location are challenging morphological deformation problems (as displayed in the left pair) in combination with staining variation and staining artifacts (as illustrated in the right pair).
Challenges of registration of histopathological images.
The challenges of registration of histopathological images include complex geometrical deformation and stain variations induced from individual data grooming steps.
The primary contribution of this study is to introduce a fully automated and robust image registration method that is able to bargain with circuitous deformation bug commonly found in biological imagery. For evaluation, three types of information are utilized, including 47 pairs of serial histopathological images, 30 light amplification by stimulated emission of radiation scanning microscope imageseighteen and 30 series ssTEM imagesxiv. The 47 pairs of serial histopathological images were collected under routine sample preparation procedures in hospitals, showing the above-mentioned deformations. The proposed method is compared with six existing paradigm registration techniques, including an elastic b-spline model for biological images (UnwarpJ)20, an improved bi-directional b-spline model for histopathological section alignment (bUnwarpJ)21, a popularly adopted image registration arroyo (SURF + affine transformation)22,23, a serial department registration approachxiv using the software package (TrakEM2)12,13,14,15, BrainAligner18 and a combination of a local contrast correction method (CLAHE)24 and bUnwarpJ. The results show that the existing image registration techniques perform poorly on the serial histopathological data with deformations and achieve less than the accuracy 70% on boilerplate while the proposed method achieves the averaged accuracy 91.49%. In alignment of laser scanning microscope brain images18, the results evidence that the proposed method is significantly better than the automatic BrainAligner18 (p < 0.001). In registration of serial ssTEM imagesxiv, the proposed method is also significantly better than TrakEM2xiv (p < 0.001).
This work combines and extends our previous efforts published in Wang and Chen25, which integrates the strengths of surface area-based and feature-based approaches and uses sparse approximation for coarse and fast global registration to greatly improve the functioning of paradigm registration. As we observed that data normalization and feature enhancement are vital for the following global characteristic-based registration and local surface area-based registration, nosotros further improve the registration method by modifying the data normalization and characteristic enhancement parts. The proposed method is able to non only reduce global and local variations among slides but also effectively identify corresponding features and with accurately identified landmark features and normalized information, the method produces valid registration outputs robust to various deformation problems ordinarily occurred in biological data, such as morphological distortions, staining variations, staining artifacts and loss of tissue.
Results
Series histopathological epitome registration
47 pairs of sequent slides were collected under routine sample training procedures in hospitals using C57 mice with IgAN (Immunoglobulin A (IgA) nephropathy), which is the most mutual glomerular disorder beyond the world26. IgAN was induced by daily injection of purified IgA anti-phosphorylcholine and pneumococcal C-polysaccharide (PnC)27 and all animal experiments were performed with the approval (permit number IACUC-11-063) by the institutional animal intendance and use Committee of the national defence medical middle, Taiwan. The tissues were fixed in 10% buffered formalin and embedded in paraffin and serial sections (4 μm) were cutting using Leica RM2155 and stained with hematoxylin and eosin (H&Due east). Images were and so captured from glass slides with standard brightfield lite microscopy (Olympus, Japan) at the magnification of 400×.
Regarding the evaluation method, as pointed out in a contempo report25 that conventional evaluation methods based on sum of squared differences (SSD) of image intensities between the target and the transformed source (meet Equation 1) to represent the registration accuracy level tin can exist misleading for tissue image applications not just because that the intensity value of the pixel in the target and the ane of the wrongly registered pixel in the transformed source may announced similar due to similarity of local tissue image features but also considering that the intensity of the pixel in the target and the one of the accurately registered pixel in the transformed source may announced different due to stain variation. Hence, the quantitative evaluation approach of the previous work25 is adopted; 5 corresponding landmarks between target images and associated transformed source images by each registration method were firstly manually marked past experienced pathologists and an automated matching system is built to compare the coordinates of the corresponding landmarks. The registration accurateness for each image pair is computed by the matching successful charge per unit over the respective landmarks (within a five pixel distance) and the operation of each registration method is evaluated by the averaged accuracies over all image pairs.
Table 1 presents the quantitative evaluation results on registration accuracies. The criterion approachesxiii,xiv,15,20,21,22 all perform poor, achieving less than lxx% accuracy on average and hence manual intervention are frequently required for accurate alignment. In comparison, the proposed method obtains registration accuracy (91.49%), greatly outperforming the benchmark methods and showing slight improvements than the author'southward previous work25. Using SPSS software28, the statistical assay results in Tabular array ii show that the proposed method is significantly improve than the criterion techniques13,14,15,20,21,22 (p < 0.01) based on the Fisher's Least Square Difference examination (LSD). Figure 3 presents the box plot of the quantitative scores, showing that the presented method often achieves higher scores than the benchmark methods. Effigy iv illustrates the quantitative evaluation results of alignment of the left image pair in Figure 1, showing that the proposed method (Fig. iv(h)) aligns the images well, achieving 100% matching score while the benchmark methods perform poor due to complex deformation problems, staining variations and disruptive local image features of biological data. Figure v shows some other image registration example by the proposed method and the criterion techniques. For the benchmark methods, the parameters adopted in the experiments are the same parameters used in the reference works14,20,21 and the parameter values are listed in the method section.
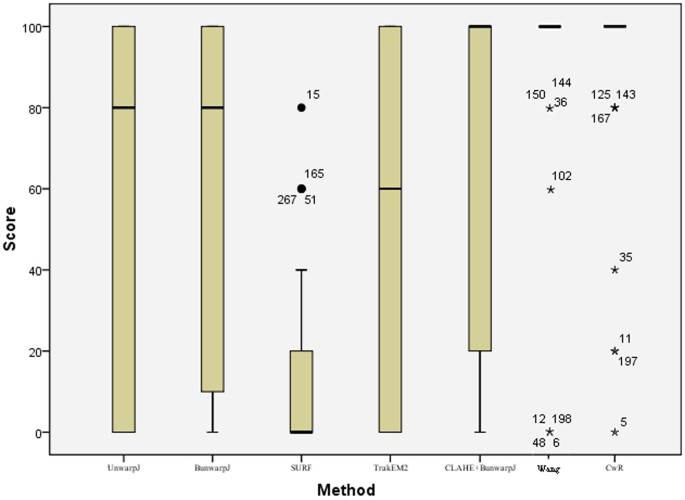
A box plot of quantitative evaluation results of histopathological image registration.
The presented method works constantly well overall and outperforms the benchmark approaches (see Table 1). Outliers > ane.v × interquartile range are marked with a dot and outliers > 3 × interquartile range are marked with an asterisk.
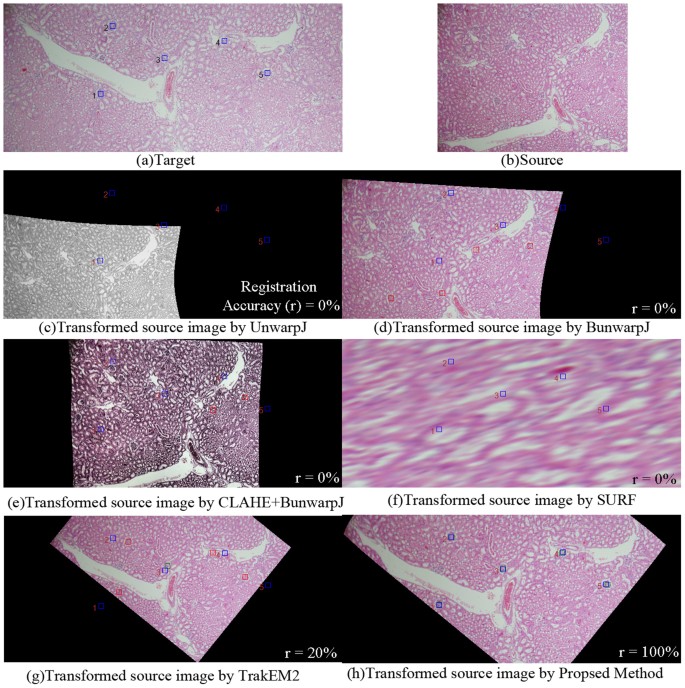
Quantitative evaluation of paradigm registration for the left image pair in Effigy i.
The blueish rectangles represent the locations of the selected landmarks defined by experienced pathologists in the target image(a); the red boxes stand for mismatches of corresponding landmarks in the transformed source image; the green boxes represent matches of corresponding landmarks in the transformed source image. In that location is no lucifer for the transformed source images by (c)UnwarpJ, (d) bUnwarpJ and (e)SURF and just one match for (f)TrakEM2. In comparison, the proposed method(grand) aligns the images well, achieving 100% target and transformed source matching successful rate.
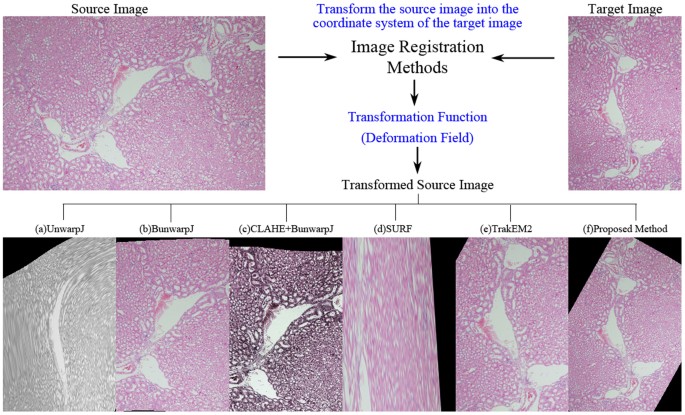
Performance comparison on registration methods.
Existing epitome registration methods perform poor on the histopathological image pair with complex distortion problems while the proposed method is able to produce a valid transformation output. (a) the transformed source image of an elastic b-spline method20 designed for biological images, (b) the output of a bi-directional b-spline approach21, (c) the transformed source image by CLAHE24 + bUnwarpJ, (d)the registration output of a popularly adopted registration method, i.e. SURF22, (due east) the output of TrakEM212,13,14,fifteen, (f) the transformed source paradigm of the proposed technique.
Laser scanning microscope images of neurons from dissimilar brains
BrainAlignereighteen is an automatic computer programme that registers pairs of 3D image stacks using landmark matching. In registration of the histopathological images, BrainAligner fails to produce valid alignment outputs. Nosotros further compare the performance of the proposed method and BrainAligner based on the data used in the BrainAligner report18. The test data are thirty light amplification by stimulated emission of radiation scanning microscope images of the Drosophila and downloaded from http://penglab.janelia.org/proj/brainaligner/BrainAligner/Download.html. Regarding the parameters used in BrainAligner, all parameters are based on the organisation default settings. For fully automated registration (without inputs of the manually marked corresponding landmarks), both global and local registration types are tested in the experiments.
Dissimilar histopathological images, local image features of laser scanning brain images appear less confusing to one another and therefore a general registration performance measurement method, i.east. the pct of pixels with similar intensity levels, is adopted to measure the registration accuracy and an automatic evaluation tool is built to conduct quantitative evaluation automatically. The registration accuracy, r, is formulated as follows.
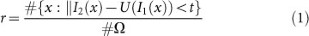
where I 1, I ii, U(I 1) represents the source, target and transformed source images; 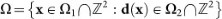 defines a mask common to the source and target images and #Ω is the size of the mask in pixels; t = 30 in our experiments.
defines a mask common to the source and target images and #Ω is the size of the mask in pixels; t = 30 in our experiments.
In our experiments, information technology is observed that with manually marked corresponding landmarks between the source and target images, semi-automatic BrainAligner performs well. However, without manually predefined landmarks, fully-automatic BrainAligner does non produce good registration outputs. In comparison, the proposed fully automatic method without manual inputs performs well. The registration accuracy scores are summarized in Table 3 and the statistical analysis of LSD test is displayed in Table 4, showing that the presented fully-automatic method is significantly better than the fully-automatic BrainAligner (p < 0.001). The notable advantage of the presented method is to save costs of manual efforts and fourth dimension in labelling landmarks. Figure vi presents the box plot of the registration accurateness scores, showing that the presented method frequently achieves loftier scores in comparing to the benchmark approaches. Figure vii displays the registration outputs of 6 paradigm pairs by the proposed fully-automatic method, the fully-automatic BrainAligner and the semi-automatic BrainAligner (with manually predefined landmarks).
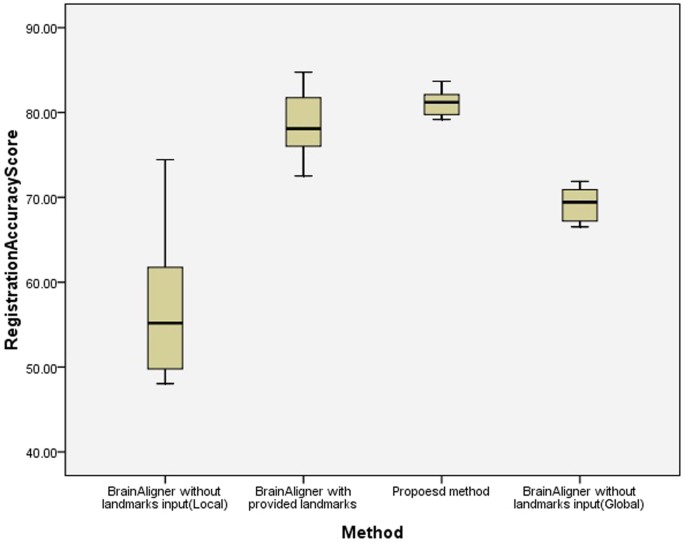
A box plot of registration accuracy scores on laser scanning brain images from BrainAligner study18.
100 indicates that the method accurately align the target and source images and 0 indicates poor registration.
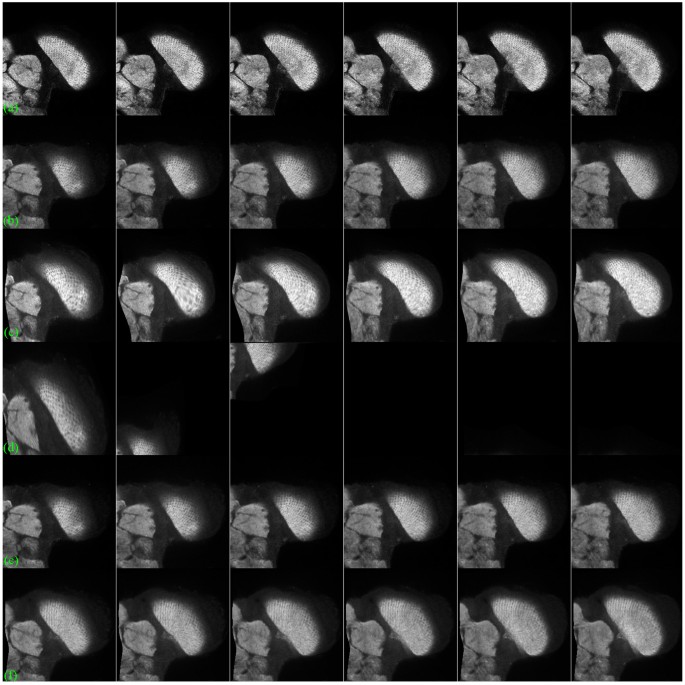
Registration of brain images by brainAligner and proposed method.
The brainAligner performs well with predefined landmarks but poor without predefined landmarks. In comparison, the proposed method without predefined landmarks produces accurate alignment outputs. (a) the target brain images, (b) the source brain images, (c) the registered source images by the proposed method (cw-R), (d) the registered source images by brainAligner without predefined landmarks (under the local registration manner), (eastward) the registered source images past brainAligner without landmarks (under the global registration way), (f) the registered source images by brainAligner with predefined landmarks.
A comparison on computational speed amid the proposed method, the semi-automatic BrainAligner and the fully-automated BrainAligner is shown in Figure 8. The processing fourth dimension for the semi-automatic BrainAligner does not include the time for the manual marker of corresponding landmarks as this information is unknown in this study. Since the manual marking time is often lengthy, therefore the processing fourth dimension by the semi-car BrainAligner might be greatly underestimated. Comparing the automatic approaches, on average the proposed method takes 3.79 second per registration while the automatic BrainAligner with the local registration way costs 22.93 second per registration on average and the automatic BrainAligner with the global registration mode costs 52.32 2d per registration on average. In statistical analysis, the presented method is significantly faster than the automatic BrainAligner approach (p < 0.001 based on Tukey HSD and LSD tests).
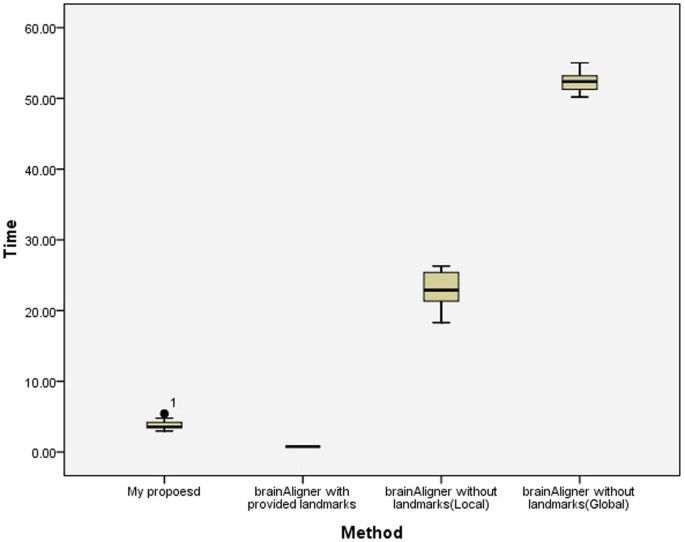
A box plot of the prototype registration processing speed on the brain images.
The processing fourth dimension (in seconds) on brain image registration past the proposed method, the semi-automatic brainAligner and the automated brainAligner. The processing time for the semi-automatic brainAligner does not include the time for manually marker corresponding landmarks as this information is unknown in this written report. Since the manual marking time is oftentimes lengthy, therefore the processing time by the semi-automatic brainAligner is profoundly underestimated. Comparing the automatic approaches, the proposed method is significantly faster than the brainAligner approach (p < 0.001 based on Tukey HSD and LSD tests) and costs 3.79 second per registration.
Serial section transmission electron microscopy (ssTEM) of drosophila neural tissue
We have besides compared the performance of the proposed method and TrakEM2 based on the data used in the TrakEM2 studyxiv. The test images are xxx serial ssTEM sections of the Drosophila first instar larva ventral nervus cord (VNC) and downloaded from http://www.ini.uzh.ch/~acardona/information.html. In evaluation, the general registration performance measurement method, i.due east. the percentage of pixels with similar intensity levels, is adopted to measure the registration accuracy (run into Equation i).
Effigy ix displays registration results of six pairs of ssTEM images by the proposed method and TrakEM2; row (a) and (b) evidence target images and source images, respectively and row (c) and (d) present registered source images past the proposed method and TrakEM2, respectively. It is observed that in this data fix, there is no distinctive morphological deformation between the target images(a) and the source images(b), but illumination variations betwixt the source and the target can be large, which tends to brand the overall registration scores lower in comparison to the BrainAligner data set. In the 2nd column of Effigy 9, it shows that TrakEM2 generates poorer alignment output(d) than the proposed method(c). Figure ten displays the box plot of the registration accurateness scores by the proposed method and TrakEM2, showing that the proposed method tends to obtain higher scores than TrakEM2. Moreover, using ANOVA assay, the proposed method is significantly better than TrakEM2 in image registration of serial ssTEM images (p < 0.001).
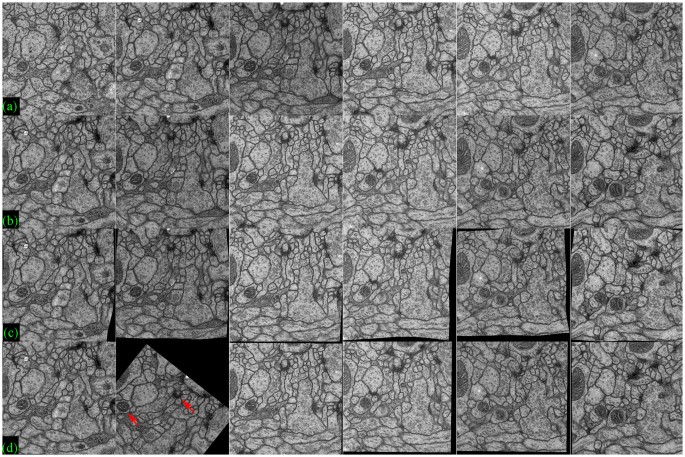
Registration of ssTEM images by TrakEM2 and the proposed method.
(a) the target images, (b) the source images, (c) the transformed source images by the proposed method (cw-R), (d) the transformed source images by TrakEM2.
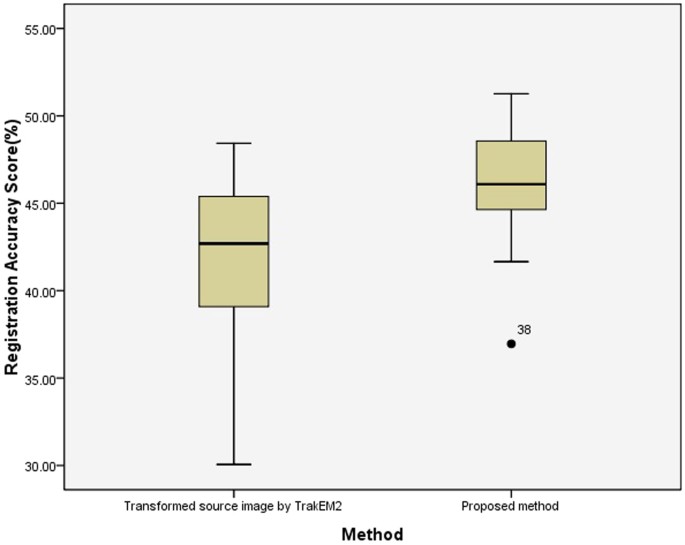
A box plot of registration accuracy scores on ssTEM images from TrakEM2 study14.
100 indicates that the method accurately align the target and source images and 0 indicates poor registration.
Discussion
We present a fully automatic elastic registration method for aligning 2d tissue images robust to morphological distortions, staining variations and staining artifacts. In addition, the presented method for detection of respective landmarks works reliably without manual intervention and the presented epitome registration algorithm is not just limited to histopathological slides merely tin likewise be applied to other anatomically or histologically divers biological images such as laser scanning microscope encephalon images and series ssTEM images. Moreover, as complex deformations are unavoidable in real life data, the presented technique will prove to exist a substantial reward for any application that requires paradigm registration. Nosotros have integrated our method into a public Java-based image processing system, ImageJ29 and the software is made publicly bachelor (http://www-o.ntust.edu.tw/~cweiwang/ImageRegistration/), assuasive the scientific community to download and use the robust prototype registration system (Cw-R).
Methods
This newspaper presents a fully automatic, robust and fast registration method that integrates the strengths of both area-based approaches and feature-based methods for biological data, containing three chief parts: (1) improved data normalization and feature extraction, (2) sparse approximation of images for coarse and fast global registration, (3) Optimize and refine local registration by area-based direct matching approach. This work combines and extends our previous efforts published in Wang and Chen25 and the comeback is made on the offset function, which is improved data normalization and enhanced feature extraction. In this section, descriptions are focused on the modified part and other parts can be constitute in the previous publication25.
Improvements on information normalization and epitome feature extraction tin can not merely help identify more than constructive respective characteristic pairs for global registration but besides heave upwards the performance of area-based local registration. Figure xi shows the results of detection of corresponding feature pairs by SURF, SIFT, the author'due south previous work25 and the proposed method, showing that (a)SURF produces poor matching, (b)SIFT generates some wrong matches and (d)the proposed method find more accurately-matched corresponding feature pairs for fast and higher level coarse registration than (c)the writer'due south previous work25.
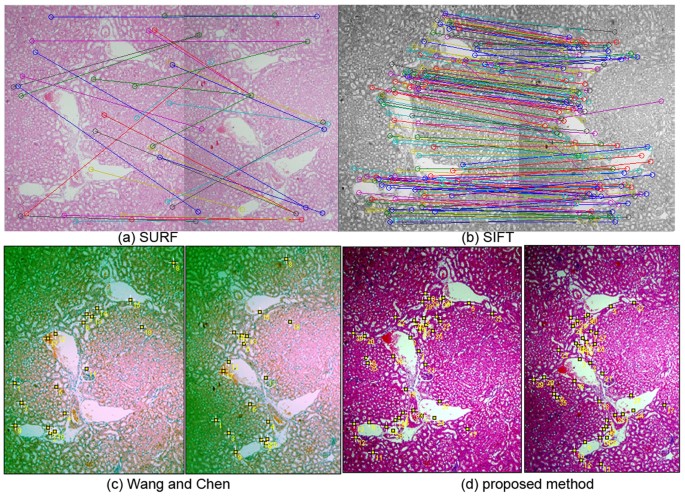
Detection of corresponding features by SURF, SIFT, the author's previous work25 and the proposed method.
(a)SURF produces poor matching, (b)SIFT generates some incorrect matches and (d)the proposed method find more accurately-matched respective feature pairs for fast and higher level coarse registration than (c)the author'south previous work.
Improved data normalization and characteristic extraction
An improved information normalization process is proposed to reduce variations on paradigm features and enhance tissue patterns. This profoundly benefits global feature matching and local area-based directing matching processes. Information technology automatically optimizes the effulgence and dissimilarity of individual color channels based on the analysis of the image histogram distribution. Furthermore, as mutual contrast stretching algorithms are sensitive to fasten, ridges or long tails bug at the tails of the histogram distribution and unable to detect the dynamic range of the main cluster, a modified method is presented here to effectively identify the dynamic range of the main cluster and deal with spike, ridges or long tails problems. Figure 12 presents (a) a raw tissue image with long tail problem, (b) the prototype generated by a commonly adopted machine contrast adjustment method (using Adobe Photoshop) and (c) the image by the presented information normalization method. Effigy 12 shows that tissue paradigm features such as nuclear patterns are greatly enhanced by the presented method.
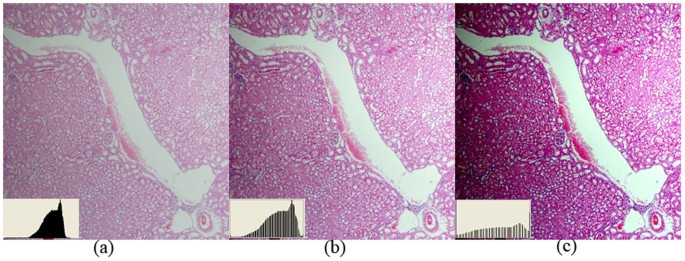
Comparing of information normalization methods.
The proposed information normalization method is able to deal with spike, ridges or long tails problems at the tails of the histogram distribution and effectively enhance tissue patterns. (a) a raw tissue image with long tail problem, (b) image by a normally adopted auto dissimilarity adjustment method (Adobe Photoshop is used here), (c) paradigm by the presented data normalization method where tissue image features such equally nuclear patterns are enhanced.
In conventional histopathological staining (H&E), Hematoxylin induces the blue staining of nuclei and Eosin induces the red/pinkish staining of cytoplasm. Based on our previous written reportthirty showing that applying histogram equalization in RGB color space performed amend in separating the nuclei from the cytoplasm than in HSL color space, both to overstate the divergence betwixt the nuclear and cytoplasmic expression in color space and to farther produce more than distinctive tissue image features, the normalization is applied to the Cherry, Greenish and Blue components of each paradigm.
In color images, the value of each pixel is represented past a vector 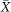 with elements the pixel values of each colour component. Assuming
with elements the pixel values of each colour component. Assuming 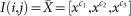 a random vector, which models the pixel value for each color component c 1, c ii, c 3 in a colour image. Firstly, the lower and the upper bound intensity levels of the histogram of each channel, x depression and x high , are computed past the equations beneath. Given a histogram distribution H, where H(10) is the number of pixels with intensity level x, the lower and the upper bound values for transformation are formulated equally follows.
a random vector, which models the pixel value for each color component c 1, c ii, c 3 in a colour image. Firstly, the lower and the upper bound intensity levels of the histogram of each channel, x depression and x high , are computed past the equations beneath. Given a histogram distribution H, where H(10) is the number of pixels with intensity level x, the lower and the upper bound values for transformation are formulated equally follows.
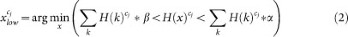

where c j : {c i, c 2, c iii}, α = 0.1 and β = 0.002.
Next, it maps the original pixel value 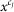 in the range from
in the range from 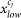 to
to  to new value
to new value 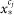 in the valid intensity scale from
in the valid intensity scale from 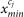 to
to 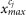 .
.

The data normalization greatly reduces stain variation and enhances tissue patterns, assisting the following characteristic extraction model to place valid respective landmarks and improving image registration outcomes. An illustration is given in Figure 13; (a) the source image, (b) the target image, (c) and (d) are the images applied with the presented data normalization method with automatically identified corresponding landmarks, (eastward) and (f) are images with deformation grid, (thou) is the deformation field and (h) is the transformed source paradigm that is well aligned to the target image. The tissue patterns of the images applied with the presented data normalization method are greatly enhanced, assisting the post-obit feature extraction model to identify valid respective landmarks and improving image registration outcomes.
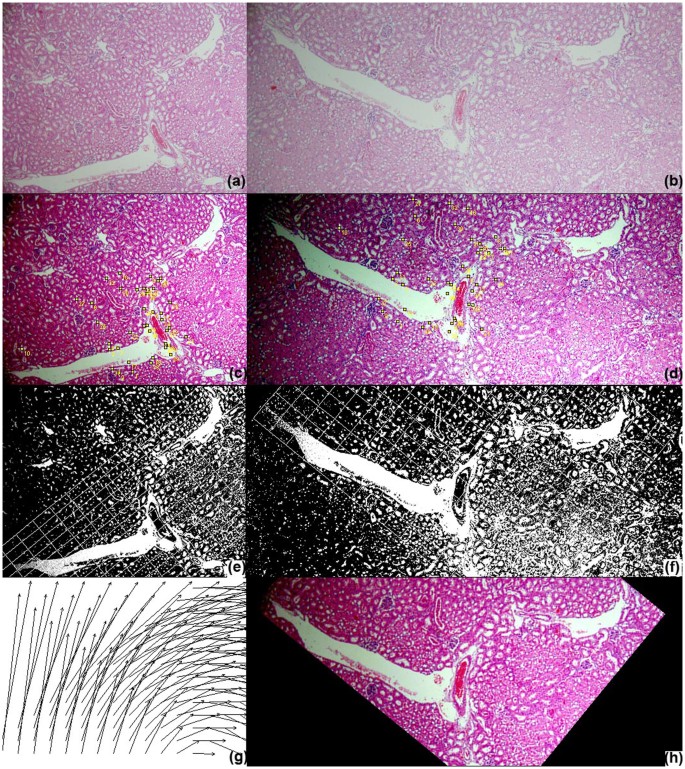
Illustration of the proposed information normalization method.
The proposed data normalization method enhances tissue patterns, reduces stain variation and assists the following identification of corresponding landmarks and image registration. (a) the source epitome, (b) the target image, (c) and (d) are the images applied with the presented data normalization method with automatically identified corresponding landmarks, (e) and (f) are images with deformation grid, (grand) is the deformation field and (h) is the transformed source paradigm that is well aligned to the target image.
For biological images, although the dyes used are visualized as having unlike colors, the resulting stains really have complex overlapping assimilation spectra. In the previous studies, colour deconvolution was used to achieve color separation in forensic image processing31 and to attain stain separation32,33 in biological image processing. Our goal is to extract the eosinophilic structures, which are generally composed of intracellular or extracellular protein, every bit prototype features for image registration and the color decomposition technique is utilized to extract independent haematoxylin and eosin stain contributions from individual histopathological images using orthonormal transformation of RGB.
In the RGB color-space, every colour is defined every bit  where r, yard, b represent the crimson, dark-green and blueish components and we can see additive color mixing as the vector addition of RGB components. To model the colors in an prototype every bit the vector addition of a desired (D) and undesired (U) components to a background color (P), new unit vectors can be defined as follows.
where r, yard, b represent the crimson, dark-green and blueish components and we can see additive color mixing as the vector addition of RGB components. To model the colors in an prototype every bit the vector addition of a desired (D) and undesired (U) components to a background color (P), new unit vectors can be defined as follows.



where  is perpendicular to
is perpendicular to  and
and  ;
; 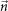 ,
,  ,
, 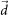 span the 3D space;
span the 3D space;  and
and 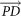 are alternative unit of measurement vectors based on the undesired and desired colors.
are alternative unit of measurement vectors based on the undesired and desired colors.
And so, colour 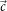 tin can be transformed to the new unit vectors.
tin can be transformed to the new unit vectors.

where 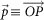 ; O is the origin in the RGB 3D space;
; O is the origin in the RGB 3D space; 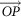 is a vector.
is a vector.
By setting u = 0, nosotros remove the undesired component and obtain the new colour 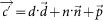 . In the case of three channels, the color arrangement tin can be described every bit a matrix of the course with every row representing a specific stain and every column representing the optical density (OD) as detected by the red, dark-green and blue aqueduct for each stain.
. In the case of three channels, the color arrangement tin can be described every bit a matrix of the course with every row representing a specific stain and every column representing the optical density (OD) as detected by the red, dark-green and blue aqueduct for each stain.

For normalization, each OD vector is divided by its total length, such that (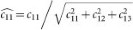 ,
, 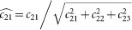 and
and 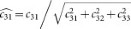 ). In this study, the normalized optical density (OD) matrix,
). In this study, the normalized optical density (OD) matrix, 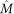 , to describe the color system for orthonormal transformation is defined as follows:
, to describe the color system for orthonormal transformation is defined as follows:

When C is the three × 1 vector for amounts of the stains at a particular pixel, the vector of OD levels detected at that pixel is equal to  . Therefore, multiplication of the OD image with the changed of OD matrix results in orthogonal representation of the stains forming the image (
. Therefore, multiplication of the OD image with the changed of OD matrix results in orthogonal representation of the stains forming the image ( ). Then, the epitome features of the red channel are extracted every bit eosinophilic structures for both loftier level feature-based coarse registration and local area-based direct matching registration.
). Then, the epitome features of the red channel are extracted every bit eosinophilic structures for both loftier level feature-based coarse registration and local area-based direct matching registration.
Parameter values used in the experiments
For the criterion methods, the parameters used in the experiments are the default parameters in the implementations of the reference works14,twenty,21 and they are the same parameters every bit used in the previous studies14,20,21. The values of the parameters for the criterion methods and the proposed method are listed below.
UnwarpJ parameter values
Initial deformation = Very Coarse, Concluding deformation = Fine, Divergence weight = 0.0, Ringlet Weight = 0.0, Landmark Weight = 0.0, Image Weight = 1.0, Cease Threshold = 0.01.
bUnwarpJ parameter values
Registration Mode = Authentic, Initial deformation = Very Coarse, Terminal deformation = Fine, Departure weight = 0.0, Curl Weight = 0.0, Landmark Weight = 0.0, Image Weight = ane.0, End Threshold = 0.01.
TrakEM2 parameter values
way = least squares (linear feature correspondences). For Calibration Invariant Interest Point Detector, initial gaussian blur = 1.60px, Steps per scale octave = 3, minimum image size = 64px, maximum epitome size = 600px. For Feature Descriptor, feature descriptor size = 8, feature descriptor orientation bins = 8, closest/next closest ratio = 0.92. Maximal alignment error = 100px, minimal inlier ratio = 0.2, minimal number of inliers = 7, expected transformation = rigid, tolerance = 0.5px. Desired transformation = Rigid, correspondence weight = 1.00. In optimization, maximal iterations = 2000, maximal plateauwidth = 200, mean factor = 3.00.
SIFT parameter values used in the proposed method
initial gaussian blur = 1.05px, Steps per scale octave = 3, minimum epitome size = 64px, maximum paradigm size = 1024px, feature descriptor size = 4, characteristic descriptor orientation bins = viii, closest/next closest ratio = 0.92, maximal alignment fault = 30px, minimal inlier ratio = 0.05, minimal number of inliers = 7, expected transformation = rigid.
Software and test images
The adult software is platform independent and thus tin can be executed in dissimilar operation systems such as Windows, Linux or Mac. The software with some test images can be downloaded from the author's website (http://www-o.ntust.edu.tw/~cweiwang/ImageRegistration/).
References
-
Mosconi, L. Brain glucose metabolism in the early on and specific diagnosis of Alzheimer's illness. FDG-PET studies in MCI and Advertisement. Eur. J. Nucl. Med. Mol. Imaging. 32, 486–510 (2005).
-
Maintz, J. B. A. & Viergever, M. A. A survey of medical image registration. Med. Image Anal. 2, one–36 (1998).
-
Cachier, P. Recalage non rigide d'images medicales volumiques - contribution aux approches iconiques et geometriques. Ph.D. Thesis, Ecole Centrale des Arts et Manufactures (2002).
-
Colina, D., Batchelor, P., Holden, K. & Hawkes, D. Medical image registration. Phy. in Med. and Biol. 3, R1–45 (2001).
-
Makela, T. et al. A review of cardiac image registration methods. IEEE Trans. Med. Imag. 21, 1011–21 (2002).
-
Oliveira, F. P. & Tavares, J. M. Medical prototype registration: a review. Comput. Methods Biomech. Biomed. Engin. 17, 79–93 (2014).
-
Zitova, B. & Flusser, J. Image registration methods: a survey. Imag. Vis. Comput. 21, 977–one thousand (2003).
-
Tan, Y., Hua, J. & Dong, G. 3D reconstruction from 2D images with hierarchical continuous simplices. Visual Comput. 23, 905–914 (2007).
-
Peng, H. et al. V3D enables real-fourth dimension 3D visualization and quantitative assay of large-calibration biological image data sets. Nat. Biotechnology 28, 348–53 (2010).
-
Pitiot, A. & Guimond, A. Geometrical regularization of deportation fields for histological image registration. Med. Prototype Anal. 12, 16–25 (2008).
-
Fiala, J. C. Reconstruct: a free editor for serial section microscopy. J. Microscopy 218, 52–61 (2005).
-
Cardona, A. et al. An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Calculator-Assisted Serial Department Electron Microscopy. PLoS Biology viii, e1000502 (2010).
-
Cardona, A. et al. TrakEM2 Software for Neural Excursion Reconstruction. PLoS ONE seven, e38011 (2012).
-
Saalfeld, South., Cardona, A., Hartenstein, V. & Tomancak, P. As-rigid-every bit-possible mosaicking and serial section registration of big ssTEM datasets. Bioinfo. 26, i57–i63 (2010).
-
Saalfeld, S., Fetter, R., Cardona, A. & Tomancak, P. Elastic volume reconstruction from serial of ultra-thin microscopy sections. Nat. Methods 9, 717–720 (2012).
-
Chakravarty, Grand. et al. The creation of a brain atlas for image guided neurosurgery using serial histological data. NeuroImage thirty, 359–376 (2006).
-
Dauguet, J. et al. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. Kidney Int. 164, 191–204 (2007).
-
Peng, H. et al. BrainAligner: 3D registration atlases of Drosophila brains. Nat. Methods viii, 493–498 (2011).
-
Alic, L. et al. Facilitating tumor functional assessment by spatially relating 3D tumor histology and in vivo MRI epitome registration arroyo. PLoS I 6, e22835 (2011).
-
Sorzano, C., Thevenaz, P. & Unser, Yard. Elastic Registration of Biological Images using Vector-Spline Regularization. IEEE Trans. Biomed. Engin. 52, 652–663 (2005).
-
Arganda-Carreras, I. et al. Consequent and Rubberband Registration of Histological Sections using Vector-Spline Regularization. LNCS, Comput. Vis. Approaches to Med. Imag. Anal. 4241, 85–95 (2006).
-
Bay, H., Ess, A., Tutelaars, T. & Gool, L. Speeded Upwards Robust Features (SURF). Comput. Vis. Imag. Understand. 110, 346–359 (2008).
-
Kroon, D. OpenSURF (including Image Warp). http://world wide web.mathworks.com/matlabcentral/fileexchange/28300-opensurf-including-image-warp (2010) Date of access:01/08/2012.
-
Saalfeld, S. CLAHE (Dissimilarity Limited Adaptive Histogram Equalization). http://rsbweb.nih.gov/ij/plugins/clahe/index.html (2009) Date of access:1/8/2012.
-
Wang, C. & Chen, H. Improved Epitome Alignment Method in application to X-ray Images and Biological Images. Bioinfo. 29, 1879–1887 (2013).
-
D'Amico, G. The commonest glomerulonephritis in the world IgA nephropathy. Q. J. Med. 65, 709–727 (1987).
-
Chao, T. G. et al. The endogenous immune response modulates the course of IgA-immune circuitous-mediated nephropathy. Kidney Int. seventy, 283–297 (2006).
-
SPSS Inc. SPSS for Windows, Rel.17.0.one. 2008. Chicago: SPSS Inc.
-
Rasband, Due west. S. ImageJ. U.S. National Institutes of Wellness, http://imagej.nih.gov/ij/ (1997–2012) Engagement of access:i/viii/2012.
-
Wang, C. & Yu, C. Automated morphological classification of lung cancer subtypes using H&Eastward tissue images. Mach. Vis. Appl. 24, 1383–1391 (2013).
-
Berger, C., Koeijer, J., Glas, Due west. & Madhuizen, H. Color Separation in Forensic Image Processing. J. Forensic Sciences 51, 100–102 (2006).
-
Ruifrok, A. C. & Johnston, D. A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23, 291–299 (2001).
-
Wang, C. Fast automatic quantitative cell replication with fluorescent live cell imaging. BMC Bioinfo. 13, ane–10 (2012).
Acknowledgements
This piece of work is jointly supported by the National Science Council of Taiwan under Grant No. NSC101-2628-East-011-006-MY3 and the Ministry of Education of Taiwan under the First-class Young Scholar Research Grant to Prof Ching-Wei Wang.
Author data
Authors and Affiliations
Contributions
C.W. designed the written report, built the method, implemented the software and wrote the paper; Due south.K. and A.C. contributed to the data collection of the histopathological data.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third political party material in this article are included in the article'due south Creative Eatables license, unless indicated otherwise in the credit line; if the material is not included under the Creative Eatables license, users will need to obtain permission from the license holder in order to reproduce the material. To view a re-create of this license, visit http://creativecommons.org/licenses/past-nc-sa/4.0/
Reprints and Permissions
About this article
Cite this article
Wang, CW., Ka, SM. & Chen, A. Robust image registration of biological microscopic images. Sci Rep four, 6050 (2014). https://doi.org/10.1038/srep06050
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/srep06050
Further reading
-
A deep learning based framework for the registration of three dimensional multi-modal medical images of the head
Scientific Reports (2021)
-
Automatic Segmentation of Bone Canals in Histological Images
Periodical of Digital Imaging (2021)
-
Fully Automatic Registration Methods for Chest X-Ray Images
Journal of Medical and Biological Technology (2021)
-
Automatic song tract landmark localization from midsagittal MRI data
Scientific Reports (2020)
-
A Robust Image Registration Interface for Large Volume Brain Atlas
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you observe something abusive or that does non comply with our terms or guidelines delight flag it as inappropriate.
How To Register And Normalize Microscope Images,
Source: https://www.nature.com/articles/srep06050
Posted by: cantyidentionevid79.blogspot.com


0 Response to "How To Register And Normalize Microscope Images"
Post a Comment